Please wait...
Microorganism Testing in Agricultural and Cannabis Products:
Detection of Four Species of Aspergillus Using Spex® 2-Color qPCR Detection Kit
| Shop Aspergillus qPCR Detection Kits Now | Have my Sales Representative Contact Me | Have a technical question? Ask Now |
As cannabis becomes legal in more and more states and countries, there has been a corresponding increase in the amount of testing, and the cannabis testing market is currently estimated to reach $1.8 trillion by 2025. Currently, 35 states have legalized its use for medicinal applications, and states continue to pass laws that allow recreational use.
Spex offers products for testing cannabinoids, terpenes, pesticides, residual solvents, mycotoxins, and heavy metals. In this technical note, we will introduce the first of a series of Quantitative Polymerase Chain Reaction (qPCR)-based kits to analyze the number of microorganisms in agricultural products.
Impurities in Agriculture and Cannabis
Agricultural products (including cannabis) around the world are routinely tested for contaminants and impurities such as heavy metal ions, pesticides, mycotoxins, and residual solvents. This is true for all agricultural products that are consumed by human beings. Heavy metals are tested by ICP-MS or ICP-OES. Residual solvents, which are small volatile impurities, are tested by gas chromatography. Pesticides and mycotoxins, which are products or pathogens like Aspergillus, are routinely tested by liquid chromatography. Many countries have specific requirements for biological contamination such as microorganisms. The presence of live microorganisms is tested in several ways. The best quantitative results are obtained by qPCR, but other methods exist, such as cell culture techniques and ELISA assays.
Methods of Testing for Microorganisms
There are many ways to detect microorganisms in agricultural products, but the most common methods include cell culture, chromogenic media, ELISA assays, and polymerase chain reaction (PCR).
Cell culture: The methodology for the detection and quantitation of microorganisms has traditionally been performed by using various forms of cell culture. Over the years, numerous culturing media have been employed that are enriched with specific concentrations of nutrient broths mixed with agar for support and grown in individual dishes/plates. The microorganisms are allowed to grow for specific durations of time (hours to days), and the direct number of colonies are counted per milliliter of fluid (CPM) or through viable cell counting to determine cell proliferation using colony forming units per milliliter of sample (CFU/mL).
Chromogenic media: In recent years, much attention has been given to the use of chromogenic media for use in plating microorganisms. This application combines the use of synthetic substrates for the detection of enzymatic microbial activities with colorimetric reactions, allowing for more specific detection of species or sub-species, often with only visual analysis. Cell culturing requires a proficient knowledge of scientific technique and requires designated lab space for laminar flow hoods, orbital shakers, incubators, and other necessary equipment. Moreover, microbiologists who are experts in these techniques are reticent to accept some of the newer methodologies, so some debate exists in cannabis and pathogen detection as to whether cell culture is indeed the “gold standard” of microbiology. Moreover, since culturing is a manual and time consuming process, the need for more rapid results has pushed the development of other assays, namely ELISA and qPCR.
ELISA assays: ELISA (enzyme-linked immunoassay) kits are enzyme-linked immunosorbent assays designed for both the rapid detection and quantitation of target proteins, antibodies, and other soluble targets of interest including cytokines, growth factors, and signaling molecules, as well as transcription factors and post-transitional gene regulators (RNAi). ELISAs are highly sensitive, with a low limit of detection, typically in the range of 0.1 ng to 0.01 ng. Their use for detecting microorganisms involves using a heat-inactivated bacterial strain or a target protein at known concentration for positive control and allows users to automate their sampling and produce far more replicates for statistical derivation in a typical 96-well format using a multi-plate reader and minimal additional equipment needs. Additionally, the reactions do not require a high degree of technical expertise, making them more suitable for use in the cannabis space.
PCR-based methods: PCR, and now the use of qPCR, has allowed for the detection and quantitation of genes found with less than 10 copies in each sample. The combined use of fluorophores also allows a researcher to quantify five genes simultaneously, with one as a reference or “housekeeping” gene. This ensures that the DNA amplified occurs in known relative concentrations to a DNA baseline, thereby minimizing human error. qPCR is slowly becoming the “go to” technology for operators in the cannabis space for microorganism detection and quantitation by virtue of such rapid generation of results and diversity of application. Despite having a reputation of being error prone, numerous studies have shown that qPCR-based microorganism detection protocols are faster and more sensitive than classical culturing/plating techniques for staph aureus, candida and E. coli (Paule et al., 2009) (Sankari et al., 2019) (Sreshtha et al., 2019). Moreover, it can be automated and routinely performed with a modest amount of equipment and technical expertise.
Microorganism Contaminants
Biological contaminants in the form of microbes are by far one of the greatest concerns for illness. In the United States, it is estimated by the Centers for Disease Control and Prevention (CDC) that 48 million people get sick from foodborne illnesses and up to 3,000 people die from foodborne diseases each year. More than 250 agents are known to cause foodborne illness and are introduced through contamination, improper handling practices and sanitation. These agents can be chemical, physical or biological.
The five types of biological agents (microbes) are bacteria, viruses, parasites, protozoa, and fungi. Fungi are a very diverse kingdom of organisms (single and multicellular) which once were considered plants. Now it is known that fungi are more closely related genetically to animals than plants.
Mold and fungi are ubiquitous in the world. Their size and spore mobility causes fungi to spread through agricultural products quickly when the environmental conditions are favorable. Significant crop losses and foodborne illness can be attributed to mold and fungi when secondary metabolites, called mycotoxins, develop. The botanicals, cannabis and food industries battle continuously with contaminants, including molds.
The more insidious toxic fungi are from the phylum Ascomycota, which includes molds, yeasts, mildews, etc. These fungi produce what are the most common secondary metabolites, called mycotoxins, associated with food contamination. Secondary metabolites are not needed for the normal life cycle of the organism. In many cases the reason for their production is unknown. One species of fungi may produce different mycotoxins and some mycotoxins are produced by multiple types of fungi. Most of the major mycotoxins of concern in human beings come from a few dozen species from the phylum Ascomycota or the sac fungi.
|
Mycotoxins |
Fungus |
Health Effect |
Source |
|
Aflatoxins: B1, B2, G1, G2 (M1) |
Aspergillus and Penicillium Species |
Hepatotoxin Carcinogen Immunosuppressant |
Cereals, Maize/Corn, Tree Nuts, Ground Nuts |
|
Ochratoxins: A, B, C, α, β, γ |
Aspergillus and Penicillium Species |
Nephrotoxin, Immunosuppressant, Renal and Hepatic Carcinogens |
Vegetables, Cereals, Vine Fruits, Wine, Beer, Coffee |
|
Trichothecenes Toxins: NT-1, NT-2, T-2, HT-2, Diacetoxyscirpenol (DAS), Nivalenol (NIS), Deoxynivalenol (DON), Fusarenon-X |
Fusarium and Gilerallazeac Species |
Hepatotoxin Carcinogen Immunosuppressant, Neurotoxin, Necrosis, Malabsorption, Skin Issues |
Grains, Rice, Oats, Cereals, Maize/Corn |
|
Fumonisins: B1, B2, B3 |
Fusarium Species |
Esophagus Cancer, Liver Cancer, Neurotoxin |
Maize/Corn, Cereals, Grains, Beer, Garlic, Beans, Asparagus |
|
Zearalenone |
Fusarium Species |
Reproductive Issues, Infertility, Early Development |
Corn, Wheat, Oats, Sorghum, Sesame Seeds, Hay, Silage |
|
Cyclopiazonic Acid |
Aspergillus and Penicillium Species |
Convulsions |
Cheese, Maize/Corn, Ground Nuts |
|
Citrinin |
Aspergillus and Penicillium Species |
Neurotoxin, Nephrotoxic Carcinogen |
Rice, Cheese, Soy, Maize/Corn, Cereals, Grains |
|
Patulin |
Aspergillus and Penicillium Species |
Neurotoxin, Mutagenic, Hepatotoxin Immunosuppressant |
Fruits, Juices, Vegetables |
|
Ergot Alkaloids |
Claviceps Species |
Digestive Disorders, Nervous Disorders, Seizures, Vomiting, Headaches |
Cereals, Grains, Maize/Corn |
As you can see in the table above, one of the most common culprits for contamination and production of mycotoxins are the Aspergillus species of fungi. While there are many species of Aspergillus, the species of most concern for the cannabis industry are Aspergillus niger, terreus, flavus, and fumigatus. In the United States, over a dozen states have requirements for testing for Aspergillus species in cannabis products. In many cases there must be no Aspergillus species detected for a product to pass due to the highly toxic nature of the mycotoxins produced by the fungus. Toxic and lethal dosages for mycotoxins can be quite small for acute poisonings. Ochratoxin A has a tolerable daily intake designated by the World Health Organization (WHO) of 5 ng/kg of body weight per day. Ochratoxin A is very toxic with an LD50 of 20–25 mg/kg of body weight. The WHO recognizes products containing more than 1 mg/kg of aflatoxins as potentially dangerous or life threatening. The Food and Drug Administration (FDA) has limits for mycotoxins for human and animal feed up to 20 µg/kg for direct human exposure.
DNA Extraction for Pathogen Analysis
It should be noted that prior to using a pathogen kit, the proper sample homogenization and DNA extraction should be performed using the Spex Genomax homogenizer and Spex DNAmax DNA extraction kits. In DNA extractions there are three basic steps which include: 1. Tissue homogenization and cell lysis; 2. DNA precipitation and extraction; 3. DNA isolation and purification.
The first step is tissue homogenization into an analytical form which reduces particle size and breaks cell walls and ruptures the nuclear envelope to facilitate the release of the genetic material. Some types of tissue require mild homogenization and lysis due to their fragile nature, while other types, such as plant material, need vigorous disruption due to their rigid cell walls. There are generally three methods for cell lysing including: mechanical or physical disruption, chemical lysis (including enzymatic lysis), and combination methods.
Physical or mechanical methods involve some type of grinding apparatus to disrupt the tissue and break apart cell walls. These methods can range from simple manual methods, including mortar and pestle, to more efficient automated laboratory homogenizers and grinders. These methods work on a variety of tissues contained in hard to digest tissues which resist cell lysis such as plant material, bone and fibrous samples.
The second step occurs when the DNA has been released from the nucleus in the cell and it becomes part of the slurry of cell materials and proteins from which it needs to be separated using chemical components and centrifugation. After the DNA has been precipitated from the cellular debris, it is then purified by either a series of washes or by the use of silica coated spin columns which bind the DNA to the column, allowing for the washing of the cellular debris from the sample.
These processes result in potentially purified genetic material that can be used in a variety of applications and processes such as polymerase chain reaction (PCR) and quantitative PCR (qPCR). These techniques often depend on the purity of and quality of the DNA extracted from sample tissue to produce accurate results. It cannot be understated that the sample processing of these tissues plays a critical role in the result. Please see our application notes on sample preparation with the Genomax and extraction using DNAmax.
Spex 2-Color Aspergillus qPCR Kit
The 2-color Aspergillus quantitative PCR (qPCR) kit is designed to detect the presence of Aspergillus niger, terreus, flavus, and fumigatus DNA in cannabis species flower, stem and/or leaf tissue(s). The Aspergillus species DNA is detected via the FAM channel, whereas the internal control is detected via the HEX channel. For machines that lack a HEX filter, VIC can be interchangeably used. Components of the kit are:
2 x Aspergillus Detection Master Mix: The master mix includes aptamer hot-start recombinant TAQ DNA polymerase (5U/µL), 0.4 mM dNTPs, primers, probes, and an inert blue dye in a PCR buffer. The PCR buffer consists of 25 mM Tris-HCl pH 8.3, 5 mM MgCl2, 62.5 mM KCl, bovine serum albumin (BSA) and PCR enhancers. The buffering system is designed for a specific and efficient amplification of Aspergillus niger, terreus, flavus, fumigatus, and internal control DNA (limit of detection is 10 copies of Aspergillus genomic DNA). Importantly, the incorporation of BSA in the master mix crowds the matrix, thereby, neutralizing PCR inhibitors that are often co-eluted with cannabis DNA. Lastly, the master mix incorporates an inert blue dye which permits easy loading of the master mix into wells of a qPCR plate.
Aspergillus Positive Control: The Aspergillus positive control template activates the FAM channel. The positive control ensures that primers and probes that are used for the detection of Aspergillus niger, terreus, flavus, and fumigatus DNA are functioning as intended.
Internal Control: The internal control activates the HEX and/or VIC channel. The internal control ensures that the template DNA can be amplified via PCR. The incorporation of internal control in qPCR is necessary to exclude false negatives that can arise from PCR inhibitors in template DNA sample.
Yellow Dye (20 x): The inert yellow dye can be added to the template DNA sample. Therefore, when the template DNA is mixed with the 2X Aspergillus Detection Master Mix, the solution turns green which provides a visual confirmation that all necessary components are present for qPCR.
DNase/RNase/Proteinase free Water: The water was purified through ultrafiltration to remove DNases, RNases, and Proteases. This water can be used to dilute template DNA or as negative control during qPCR.
ROX: Rox is a passive reference dye that is used by low ROX and high ROX qPCR machines for well-to-well signal normalization.
Spex Aspergillus Kit Protocol
Once all components in the kit are thawed in ice, the tubes are mixed by inversion to ensure homogeneity. Before first use, ROX is added to the 2X Aspergillus Detection Master Mix to ensure qPCR machine compatibility. Afterwards, 10 µL of the 2X Aspergillus Detection Master Mix is added to designated wells of a qPCR plate. Then, in a separate 1.5 mL tube the following components are mixed:
|
Component |
Volume (10 µL) |
|
20X Yellow Dye |
1 µL |
|
DNA Template* |
1-6 µL |
|
Internal Control Template |
2 µL |
|
Water |
up to 10 µL |
* For positive control replace DNA template with 2 µL of Aspergillus Positive Control Template. For negative control, do not add any DNA template.
Lastly, after a brief centrifugation, the components are pipetted to wells containing the 2X Aspergillus Detection Master Mix and qPCR is performed using the following conditions:
|
Step |
Temperature (°C) (10 µL) |
Time (10 µL) |
Number of Cycles |
|
Denature |
95 |
1 min |
1 |
|
qPCR Detection |
95 |
10 sec |
35 |
|
60 |
30 sec |
Results
Results from the qPCR should be available within 55 minutes, depending on the qPCR machine being used. The figure below demonstrates results that can be expected when using the 2-color Aspergillus qPCR kit.
Figure 1A
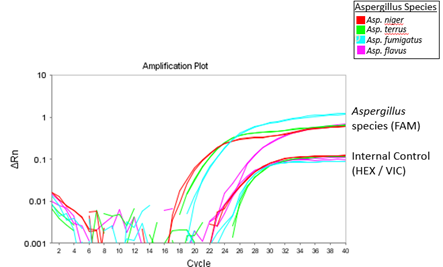
Figure 1B
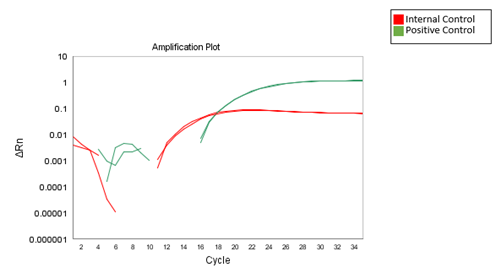
Figure 1. Expected results when using the 2-color Aspergillus qPCR kit. A. The 2X Aspergillus Detection Master Mix was used to amplify Aspergillus niger, terreus, flavus, and fumigatus genomic DNA. The amplification of Aspergillus species genomic DNA and internal control template was detected via the FAM and HEX channel, respectively. B. Amplification curves of positive and internal control template when using the 2X Aspergillus Detection Master Mix.
Conclusions
In this article, we presented a new Spex 2-Color Aspergillus qPCR Kit for measuring the total amount of Aspergillus in a sample of cannabis. The kit includes 4 different strains of this pathogen with the four strains labeled with a FAM tag. A second color is used to measure the internal standard on the HEX/VIC channel.
The PCR kit produces consistent and accurate data regarding the presence of Aspergillus contamination in agricultural and cannabis products using both positive and negative controls. These results are more consistent than alternative colony plating methods for pathogens using standard cell culture techniques, in which microorganisms may be killed during sample preparation which then prohibits the accurate growth of the pathogen resulting in falsely low or negative results.
This kit was produced in response to the increasing number of states requiring quantitative proof that the cannabis samples do not contain dangerous amounts of several targeted microorganisms like bacteria (E. coli), viruses (Hepatitis) and other fungi. We plan to introduce additional kits that measure other microorganisms of interest including E.coli, Salmonella and total yeast and mold.
| Download the complete technical note |